
The Dairy Centre launches research into the microbiome of milk and dairy derivatives
Scientific developments and technological innovations have radically altered the field of research into microorganism ecology. Molecular methods are now able to identify microorganisms that are impossible to detect with culture-based techniques, providing an enormous boost to the field of metagenomics. Next generation sequencing (NGS) enables to study the bacteria, fungi, and yeasts from different environments, leading to significant food research using this technique in recent years. Microorganisms are an essential part of the various fermentation processes involved in the preparation of many foods, and this technique is highly suited to identifying them. It is also a technique that is extremely helpful in ensuring food safety, since it provides detailed information on pathogenic microorganisms.
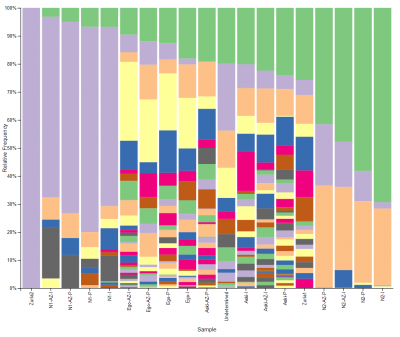
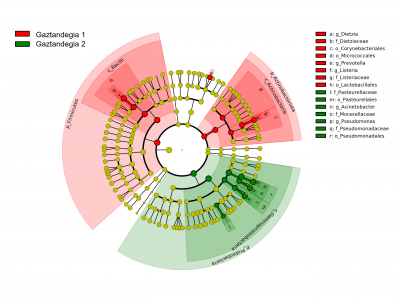
At the Dairy Centre we are implementing the iSeq 100 sequencer from Illumina to expand our knowledge of dairy microorganisms and broaden the range of services we offer to our customers. This compact desktop sequencer can identify microorganisms from different samples, even reaching specie or strain levels. This makes possible to gather information on the communities of microorganisms in raw milk, and therefore on animal health, the milking system and the efficiency of the L+D processes. Furthermore, it provides information about the microorganisms involved in the fermentation process in the cheese and dairy industry that can jeopardize the viability of the final product (whether they are technologically relevant or not). Besides, the technique can prevent problems during food preparation that lead to defective products that cannot be sold and avoid economic losses and food waste. In addition, since the food will be monitored from start to finish, the study will have a direct impact on consumer safety.

The development of this technique is a major step in the research on milk and dairy products’ microbiome. Beyond information on the entire microbiome, it offers the opportunity to go deeper into the research of each of the selected microorganisms. In general, it will make it possible to broaden our knowledge of these species in both cheese and dairy products, which are particularly relevant for a variety of reasons (from a sanitary point of view, because they are involved in the fermentation process, because they influence cheese affinage, etc.).
The information that will be gained from this development will provide added value to the milk and dairy sector of the Basque Autonomous Community.

